Current Focus
Our main aim is to ensure the safe integration of cutting-edge genomics technologies into the transfusion medicine laboratory.
The consortium members have agreed on a 2-year R&D project consisting of 10 work packages. The details of the work packages and how the project is directed is regulated in the Consortium and Members’ Agreement, which was signed by the members on the 16th April 2021.
Aims of the R&D project- To deliver an affordable Applied Biosystems™ DNA array research test for the typing of donors and patients for Human Erythroid Antigens (Blood Groups), Human Platelet Antigens (HPA) and Human Leukocyte Antigens (HLA)
- Other markers relevant to transfusion and transplantation
- To explore regulatory approval for the array in the different territories
Key objectives of the R&D project- To deliver the Pre-Clinical Study II by December 2021
- To deliver the Clinical Study in 2022
- To use the results of both studies to seek regulatory approval for the arrays we have developed
The consortium's project timeline is displayed below. You can click the header of each section to expand it and see more detailed information about each project phase.
-
2017 - The first years
In 2017 the Founding Members of the Consortium, being Cambridge University Hospitals, NHS Blood and Transplant, Sanquin, the New York Blood Center, Brigham and Women’s Hospital and Thermo Fisher Scientific started their joint R&D activities.
In their first large-scale study, named Pre-Clinical Study I, it was investigated whether the Applied Biosystems™ Axiom™ UK Biobank (UKBB) array (Astle et al., Cell, 2016) could be rendered suitable for the typing of Human Erythroid Antigens [HEA], Human Platelet Antigens (HPA) and Human Leukocyte Antigens (HLA).
With samples and antigen typing data from nearly 8,000 blood donors it was shown that nearly all clinically relevant HEA and HPA antigens can be typed accurately with the UKBBv2 research array. With HLA class I and class II types, at 2-field resolution, also being inferred from the genotyping results by imputation at high accuracy (Gleadall et al., Blood Advances, 2020).
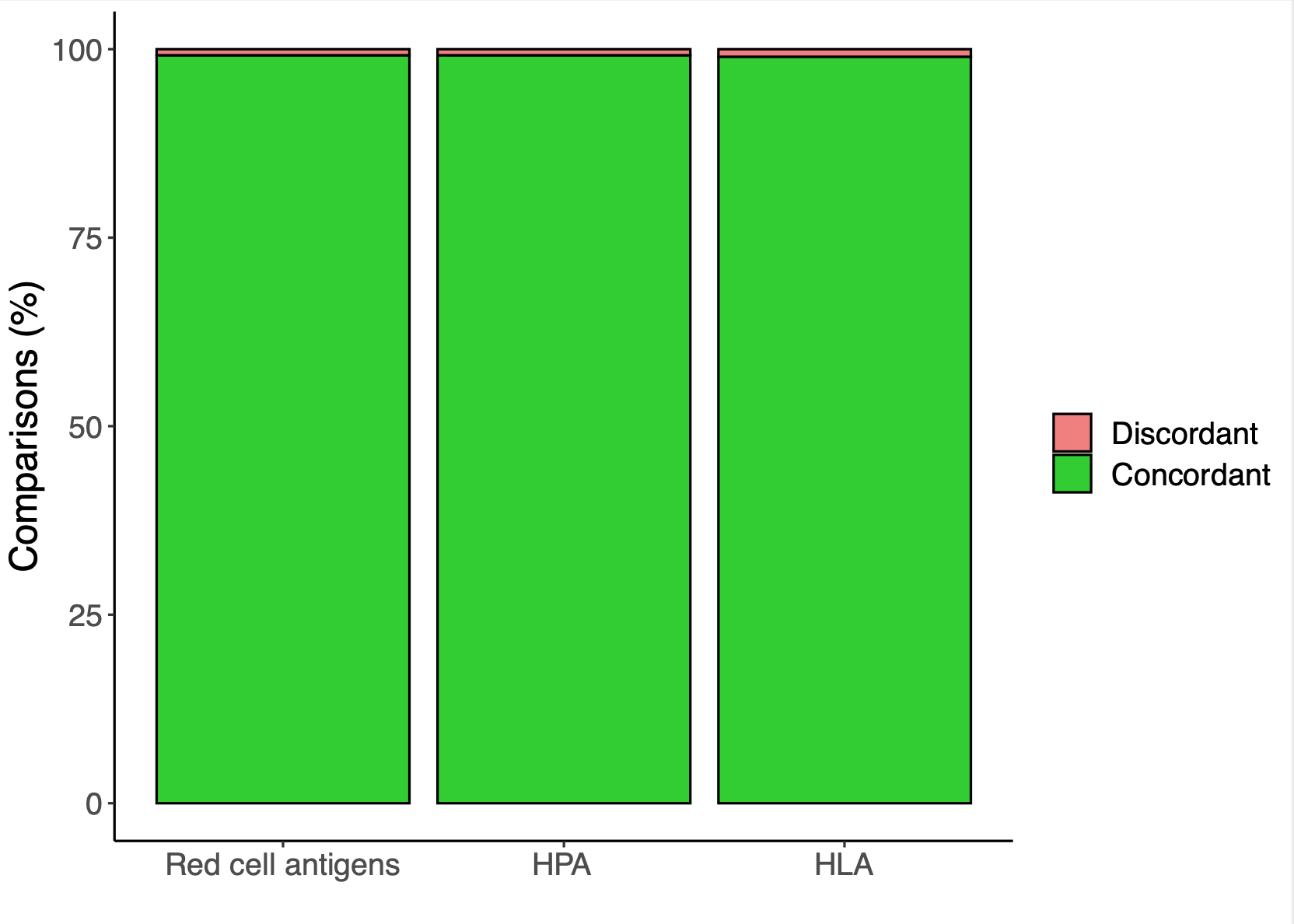
Antigen typing accuracy achieved during Pre-ClinicalStudy I. -
2019 - New members join
Based on the exciting results of the Pre-Clinical Study I, the national blood services of Australia, Canada, Finland and South Africa decided to join the Consortium in 2019.
The 10 member organisations of the Consortium then devised a 2-yr R&D plan consisting of a Pre-Clinical Study II and Clinical Study.
During these two studies, the goal was for the next versions of the UKBB array (UKBB v2.2) and a new array tailored to the needs of blood services to be designed and accredited.
-
2020 - Further Array Development
The UKBB v2 array and its preceding versions contains probesets for 800,000 DNA variants (Bycroft et al., Nature, 2018.).
These arrays were designed for large-scale population genome wide association studies (GWAS). Donors from several blood services have provided consent for being included in such GWAS efforts, with the primary purpose of these studies being the unravelling of the genetic architecture of common diseases and medically relevant traits (Vuckovic et al., Cell, 2020.).
However, blood services and hospitals may prefer to type donors and patients only for DNA variants which are important for selecting the most suitably matched units of blood and platelets for patients requiring regular transfusions.
To meet this need the consortium set out to develop the Applied Biosystems™ Axiom™ Universal Blood Donor Typing (UBDT PC1) array, which can type 50,000 DNA variants and only content relevant for blood typing alongside an improved version of the UKBB array, the UKBB v2.2 array.
-
2021 - Pre-Clinical Study II
In order to evaluate the performance of the newly developed UBDT and UKBBv2.2 research arrays, the consortium members assembled a panel of DNA samples from approximately 7,000 blood donors, with each blood service contributing a proportion of samples corresponding to the size of the population it serves.
For the study DNA samples and the accompanying antigen typing data and relevant metadata were maintained by Cambridge University Hospitals for use in the approved studies.
To evaluate the arrays GENETITAN-MC instruments were installed at the laboratories of three of the Consortium’s member organisations, being NHS Blood and Transplant (H&I laboratory, NHSBT Colindale, London, UK), the New York Blood Center (the National Genomics Laboratory, Kansas City, Missouri) and Sanquin (Amsterdam, the Netherlands).
The 7,000 Pre-Clinical Study II DNA samples were genotyped at these locations in triplicate, while the latter two laboratories evaluated the UBDT PC1 array, and the UKBB V2.2 array was evaluated by the NHSBT laboratory in London.
Following genotyping, the data was stored within the BGC Safe Research Computing Platform (SRCP) provided by the High Performance Computing Service at the University of Cambridge.
Dr. Nicholas Gleadall from Cambridge University Hospitals and NHS Blood and Transplant, Dr Bill Lane from The Brigham and Women’s Hospital and Dr Jeremy Gollub from Thermo Fisher Scientific continued to collaborate to perform concordance analysis at both genotype and antigen level. They then used the results of this analysis to fine tune and improve the array design ahead of the Clinical Study in 2022.

Overall Pre-Clinical Study II workflow We also planned in late 2021 to install a GENETITAN-MC instrument in the Lifeblood centre in Brisbane, Australia, to perform an evaluation experiment.
-
2022 - Clinical Study
Following a final round of array design, informed by the results of Pre-Clinical Study II, the consortium proceeded with the Clinical Study.
In this study the UBDT and UKBB research arrays were challenged with another set of DNA samples, again from approximately 7,000 blood donors.
We used the results of this study to engage with appropriate regulators to seek accreditation for the developed arrays.
This study was delivered by the end of 2022.
-
2023 - Preparing for launch
In partnership with Cambridge University and the UK's NIHR BioResource, NHSBT typed 92,000 blood donors using the STRIDES (STRategies to Improve Donor ExperienceS) BioResource.
NHS England made funding available for the new test - the Axiom Total Blood Typing Solution DNA test - to be made available for all patients with sickle cell disorder, thalassemia and other inherited anaemias. The test will improve the accuracy of matching for the many blood groups, improving the quality of transfusion care for people who need regular transfusions.
-
2024 - Market entry and standards development
It is expected that the Axiom™ Universal Blood Donor Typing (UBDT) array will be made available in April 2024, initially for research use only.
The consortium will continue our educational programme and focus on the development and introduction of international standards regarding the deployment of arrays in research and clinical settings.